Have you been wondering when we’ll get some of that next-gen gene expression in human tissues up as tracks in the browser? The GNF Atlas microarray tracks are so 2004… Yes, we do have RNA-seq from ENCODE cell lines, but you can get only so far with cell lines (are they even human?). Well, wait no longer! Once we learned what the GTEx folks are up to – RNA-seq and genotyping of samples from 53 tissues in many hundreds of donors – we just had to get on board! Read on for details…
The NIH Genotype-Tissue Expression (GTEx) project was created to establish a sample and data resource for studies on the relationship between genetic variation and gene expression in multiple human tissues. In April this year the Genome Browser released the GTEx Gene Expression track, which showcases data from the GTEx midpoint milestone data release (V6, October 2015) – 8555 tissue samples obtained from 570 adult postmortem individuals. The track shows median expression level per tissue at each gene via a new bar graph display:
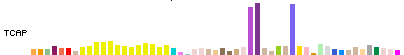
The height of each bar represents the median expression level across all samples for a tissue, and the bar color indicates the tissue (we are using GTEx publication color conventions). You can see the gene description and tissue name with expression level when you mouseover, and can view the tissue legend in glorious detail on the track configuration page. Above, notice the 3 highly expressed tissues for TCAP protein (titin-cap, used in muscle assembly) – unsurprisingly in this case, heart (2 sub-tissues) and skeletal muscle.
In the tissue mix sampled by GTEx, you’ll find a dozen brain sub-tissues, a handful of cardiovascular tissues, and bits from digestive, reproductive, and endocrine systems. For a nice summary of the tissues assayed, check out the GTEx project portal. Not so interested in all the tissues? Turn on the tissue filter and limit the graph to show just your faves!
Once you’ve found your favorite gene, you can drill down for more detail. A nice boxplot showing the range for all samples and the sample count is right here on the details page:

You’ll also see this plot on the new RNA-Seq Expression panel of the UCSC Genes detail page:
If gene-level calls aren’t your thing – you’re more of a deep diver and want to see the actual RNA-seq coverage – you might find the newly released GTEx Signal Hub just your style. We were fortunate to be able to team up with the Global Alliance crowd here within the UCSC Genomics Institute and convince them to pump all the available GTEx RNA-seq through their hot new Toil pipeline (along with twice as much cancer data) to produce signal graphs. A round of ‘biggification’, lifting and track configuration (gotta have those GTEx colors!) produced the hub. Find it on the Public Hubs panel of the Track Hubs page, which you can navigate to via the My Data > Track Hubs menu option in the top blue bar.
Did I mention you can find the GTEx gene track and the GTEx Signal hub on both the hg19 (GRCh36) and hg38 (GRCh37) genome browsers?
Give the new tracks a spin! To get you started, here’s a session:
Now enjoy!!
If after reading this blog post you have any public questions, please email genome@soe.ucsc.edu. All messages sent to that address are archived on a publicly accessible forum. If your question includes sensitive data, you may send it instead to genome-www@soe.ucsc.edu.



Hi Kate, did a little digging around trying to learn more about the RNA info on the browser, and found your blog! Looking forward to further insights!
Hi Kate,
Any chance you can point to the studies regarding how DNA can cause variation in keratin nail plate?
Thank you!
Hi,
Great insights. I am focused for my PhD work on GTEx and look forward to learning more.